The Tibetan wheat sequencing project
Tibetan wheat is grown under environmental constraints at high-altitude conditions, but its underlying adaptation mechanism remains unknown. Here, we present a draft genome sequence of a Tibetan semi-wild wheat accession Zang1817 and re-sequence 245 wheat accessions, including world-wide wheat landraces, cultivars as well as Tibetan landraces. We demonstrate that high-altitude environments can trigger extensive reshaping of wheat genomes, and also uncover that Tibetan wheat accessions accumulate high-altitude adapted haplotypes of related genes in response to harsh environmental constraints.
This project was performed by Wheat Genetics and Genomics Center (WGGC) at China Argiculture University (CAU).
Section 1: Zang1817 genome assembly
The Zang1817 genome assembly reported here is a sequenced genome of a semi-wild form of hexaploid wheat to date.
First, we generated more than 240× coverage sequencing reads from a series of PCR-free Illumina paired-end libraries and mate-paired libraries. The contig and scaffold assembly was performed using the DeNovoMAGIC3 software platform, and additional Chromium 10X genomic data were utilized to support scaffold validation and further elongation. Finally, 384,307 scaffolds with N50 of 37.62 Mb were generated, and formed the basis of a total assembly size of 14.71 Gb for Zang1817. The assembled scaffolds were ordered into chromosomes with the aid of the Chinese Spring (CS) IWGSCv1 reference genome.
High-confidence/Low-confidence protein-coding genes and transposable elements were further annotated.
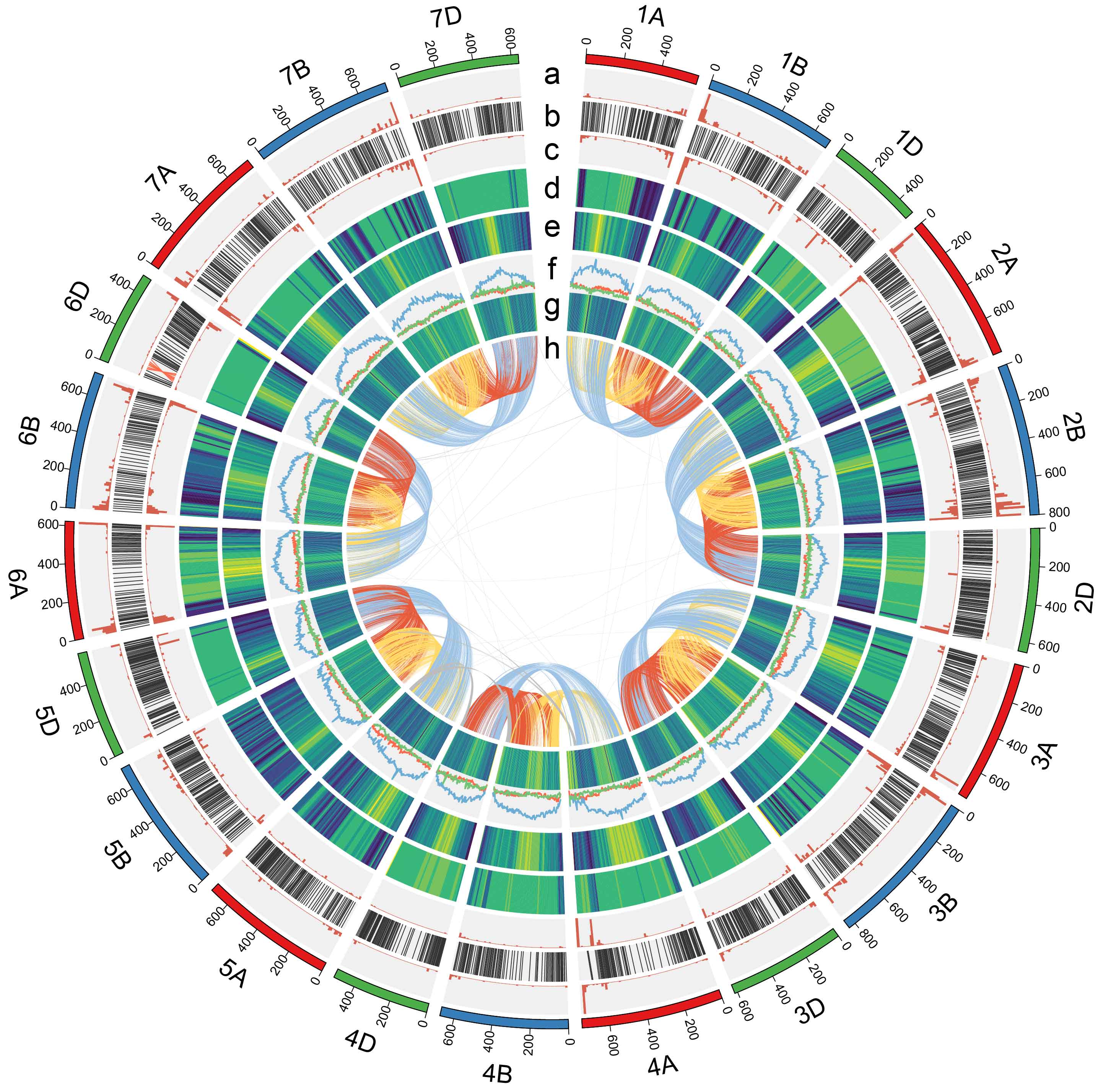
Concentric circle diagram shows distribution of the genomic features of Zang1817
Resouces available: Zang1817 genome assembly and annotation
Zang1817 genome assembly has been deposited at DDBJ/ENA/GenBank under the accession JACSIS000000000.
High-confidence gene model (GFF3): Zang1817_PseudoGenome_HC_gene_model.gff3.gz [download].
Low-confidence gene model (GFF3): Zang1817_PseudoGenome_LC_gene_model.gff3.gz [download].
Function annotation of gene model (TSV): Zang1817_PseudoGenome_Function_Annotation.txt.gz [download].
Transposable element annotation (GFF3): Zang1817_TE.gff3.gz [download].
Section 2: Whole genome resequencing
The re-sequenced collection contains 308 hexaploid wheat accessions in total, covering 74 Tibetan semi-wild accessions, 43 Chinese cultivars accessions and 102 Chinese landrace accessions (including 35 Tibetan landrace accessions), and 89 accessions from countries word-widely beyond China. The re-sequenced 245 wheat accessions in this study and previous published 63 accessions were combined together in this study.
Resouces available: sample metadata
Metadata of Resequencing samples in this study (XLSX): Sample_metadata.xlsx [download].
Metadata of Resequencing samples in this study (XLSX) in ㊥: 样本信息.xlsx [download].
Online metadata query form
Resouces available: resequencing data
The genomic sequencing reads of 245 sample in this project have been deposited to NCBI Sequence Read Archive with accession PRJNA596843. These data are also available at National Genomics Data Center (NGDC) hosted by Beijing Institute of Genomics (BIG) with BioProject ID: PRJCA002215.
Resouces available: genomic variation dataset (powered by SnpHub)
Genomic variation could be retrived and analyzed on SnpHub, access link.
Citation
Weilong Guo*, Mingming Xin*, Zihao Wang*, Yingyin Yao, Zhaorong Hu, Wanjun Song, Kuohai Yu, Yongming Chen, Xiaobo Wang, Panfeng Guan, Rudi Appels, Huiru Peng#, Zhongfu Ni#, Qixin Sun#. (2020). Origin and adaptation to high altitude of Tibetan semi-wild wheat, Nature Communications, doi:10.1038/s41467-020-18738-5.
*These authors contributed equally to this work.
#Correspondence and requests for materials should be addressed to Huiru Peng (email: penghuiru at cau.edu.cn), Zhongfu Ni (email: nizf at cau.edu.cn) or Qixin Sun (email: qxsun at cau.edu.cn)
External links
•Triticeae Multi-omics Center:
The Triticeae Multi-omics Center holds and collects data for species in Triticeae tribe. Some useful utilities, such as
- Genome BLAST,
- Sequence retrieval,
- Genome browser
and others are available for Zang1817.
•Triticeae-GeneTribe (TGT)
Collinearity-incorporating homology database for Triticeae genomes. Zang1817 genome has been incorporated into TGT and is ready for analysis.
Quick examples:
- Finding homologues between Zang1817 and Chinese Spring (IWGSC RefSeqv1.1),
- Macro collinearity of chr1A between Zang1817 and Chinese Spring (IWGSC RefSeqv1.1).

