Database / Webserver

Wheat-SnpHub-Portal
Genomic variation datasets of wheat and its progenitors
(GigaScience, 2020)

WheatUnion
Wheat Resequencing Variants joint Database / Webserver
(2020, Collaboration)

WheatCompDB
Genetic Relationship Information Database / Webserver for Wheat Germplasms
(Plant Physiology, 2022)

Triticeae-GeneTribe
Connect Emerging Assemblies in Triticeae by Genes
(Molecular Plant, 2020)
wGRN
A platform using wheat integrative regulatory networks to guide functional gene
discovery
(Molecular Plant, 2023)

wLNCdb
Wheat long non-coding RNA (lncRNA) Database / Webserver
(Seed Biology, 2023)
pNOGmap
A Database / Webserver for nucleus organellar genes in Poaceae
(Plant Physiology, 2023)

WheatCNVb
A Database / Webserver for CNV block profiling and variety comparison
(Genome Biology, 2024)
DualPE-Finder
Plant genome editing tools for deletions, replacements and inversions
(Nature Plants, 2024)
Software

SnpHub
An easy-to-set-up web server framework for exploring large-scale genomic variation data in the
post-genomic era with applications in wheat
(GigaScience, 2020)

ggComp
A pairwised comparison method to identify similar genetic regions and shared CNV regions
(Plant
Physiology, 2020)

IntroBlocker
Semi-supervised algorithm for ancestral genomic blocks dissection
(Nature Communications, 2022)

GeneTribe
A tool for performing collinearity-incorporating homology inference
(Molecular Plant, 2020)

IGTminer
A tool for identifying the evolutionary trajectories of nuclear organellar genes
(Plant
Physiology, 2023)

PanSK
K-mer based approach for identifying PAV and haployptes of wheat SSP genes at pangenome level
(Molecular Plant, 2024)

LcQTH
Toolkit for rapid quantitative trait mapping with ultra-low-coverage sequencing
(Plant Communications, 2024)
Dataset
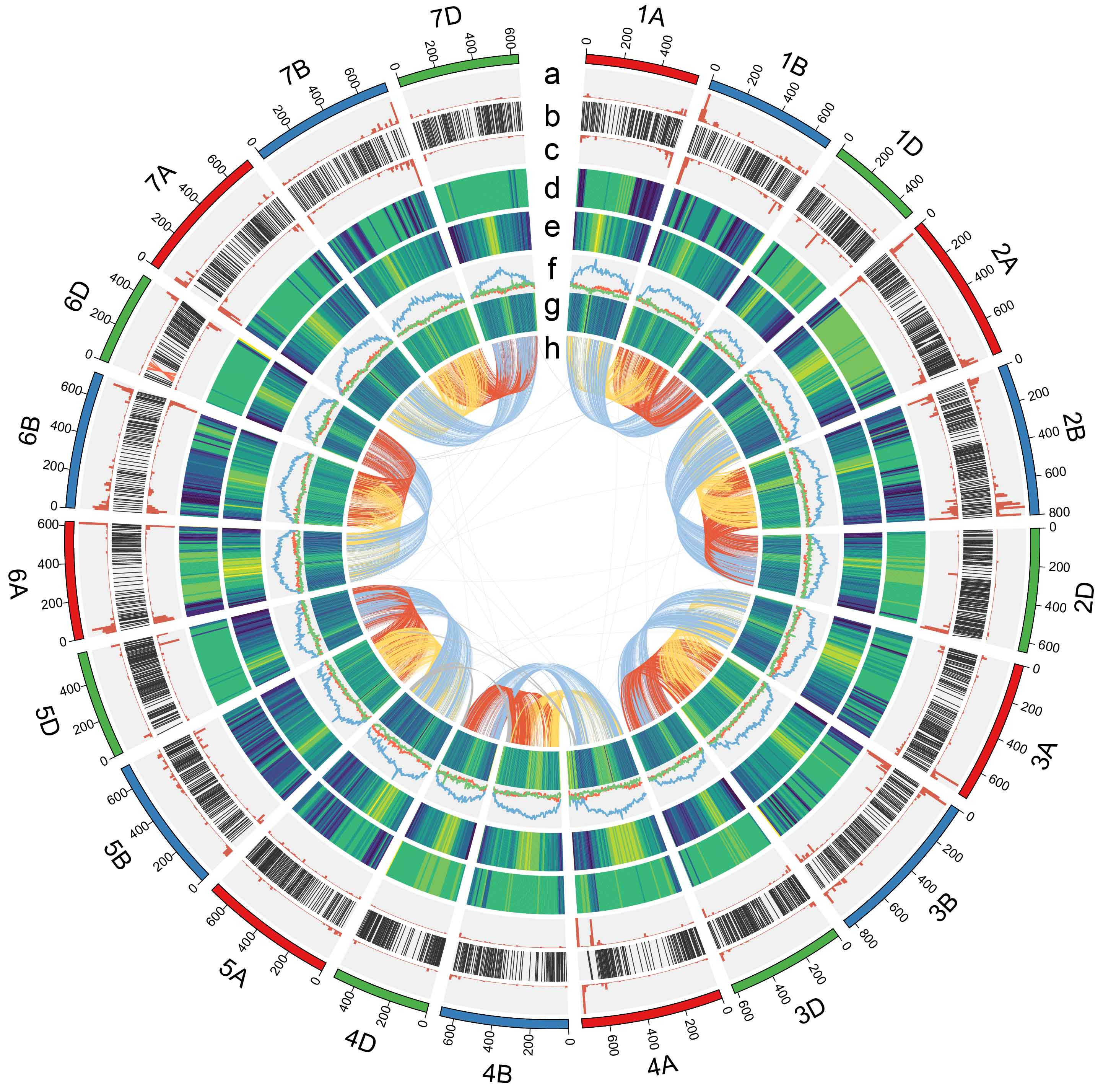
The Tibetan wheat sequencing project
Zang1817 genome assembly and Whole genome resequencing
(Nature Communications, 2020)
Grain Translatome Browser
A browser for analyzing the translatomic data of developing wheat grains
(Plant cell, 2023)

Chinese Spring genome near-complete assembly (CS-CAU)
Chinese Spring genome near-complete assembly (CS-CAU)
(Molecular Plant, 2025)
