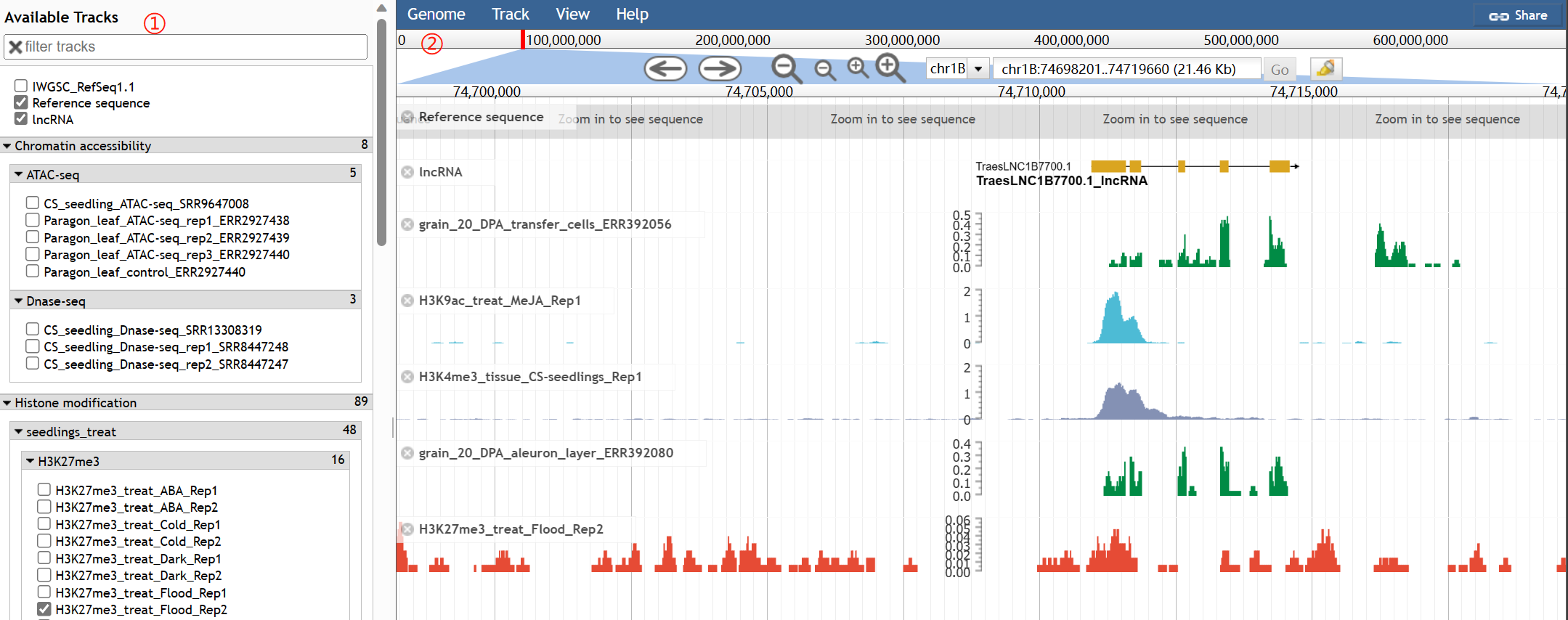
Using this tool, you can browse multi-omics data track of interested gene. Notably, we also implemented the wGRN_v1 annotation for users to screen interested gene models. In this example, wGRN_v1 corrected CDS and added UTRs of TraesCS1A03G0652000. The JBrowse tool ( https://jbrowse.org/jbrowse1.html) was used here.
1. In the left panel, you can select tracks of interest, including chromatin accessibility (ATAC-seq/DNase-seq), histone modification (H3K27me3, H3K4me3, and H3K9ac ChIP-seq), and expression levels (RNA-seq). STAR and BWA were used to align sequencing data to the genome.
2. Right panel shows selected tracks.