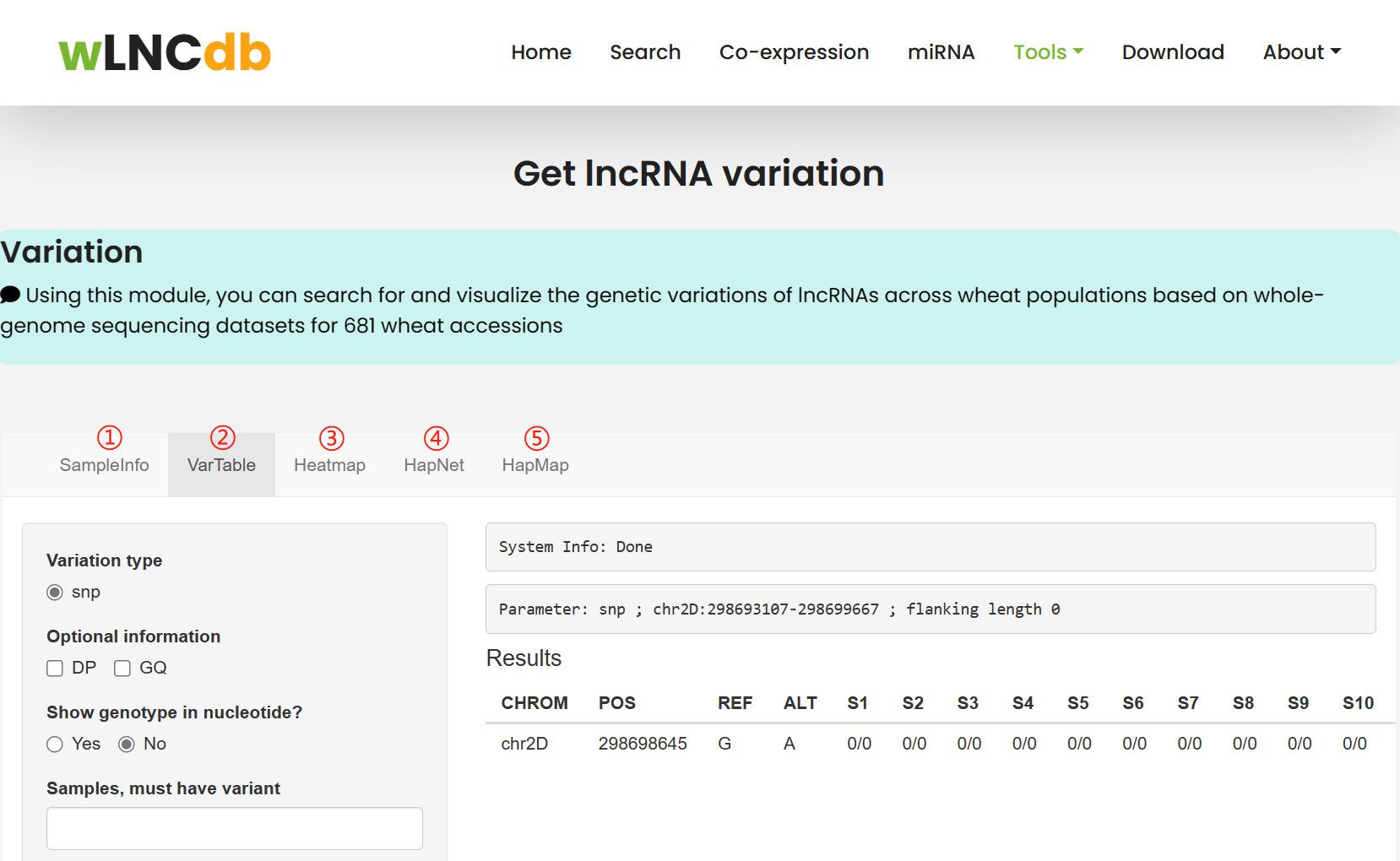
Using this tool, you can submit lncRNA position to query genetic variation of lncRNA. The SnpHub tool ( http://guoweilong.github.io/SnpHub/) was used here.
1. providing the sample description information for the data set.
2. users can query region-specific SNP/INDEL tables for a list of samples. To be consistent with the VCF format, the exported genotypes are denoted as 0/0, 1/1, 0/1 or ./. .
3. provides a way to visualize genotype variations across multiple samples in an intuitive interface.
4. Haplotype networks represent the relationships among the different haploid genotypes observed from the sample list
5. The HapMap channel provides a way to project the allele distribution of a single site geographically on a map, utilizing the provided resource-gathering locations.